Generating bit patterns
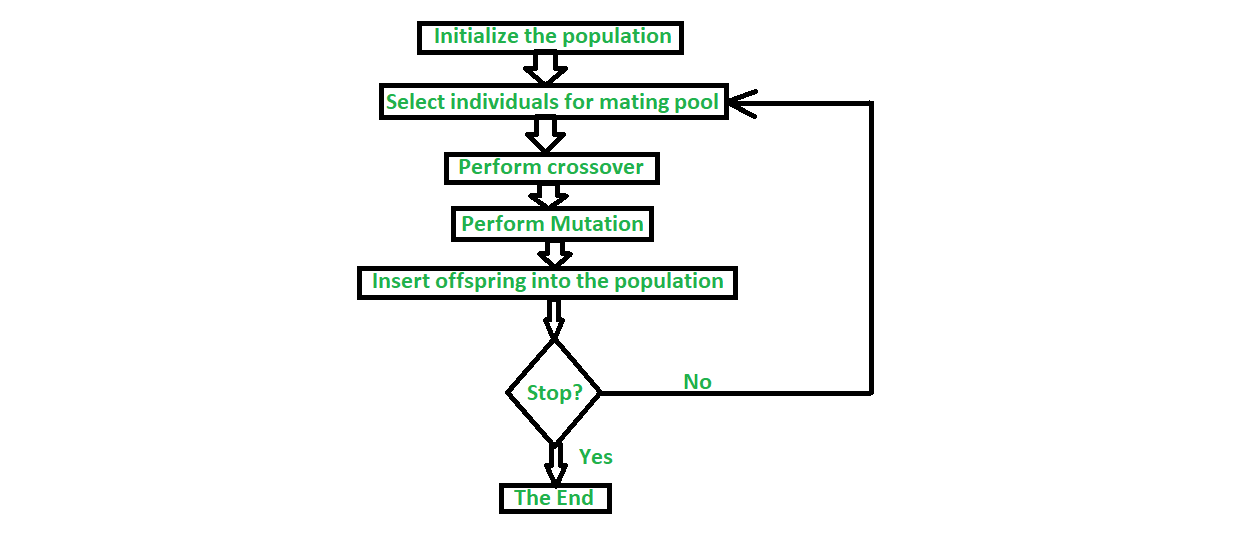
The following example shows you how to generate a bit string that would contain 15 ones, based on the One Max problem.
import random
from deap import base, creator, toolsDefine the evaluation function. It is the first step to create a genetic algorithm.
def eval_func(individual):
target_sum = 15return len(individual) - abs(sum(individual) - target_sum),
Now, create the toolbox with the right parameters:
def create_toolbox(num_bits):
creator.create("FitnessMax", base.Fitness, weights=(1.0,))creator.create("Individual", list, fitness=creator.FitnessMax)
Initialize the toolbox:
toolbox = base.Toolbox()
toolbox.register("attr_bool", random.randint, 0, 1)
toolbox.register("individual", tools.initRepeat, creator.Individual,
toolbox.attr_bool, num_bits)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
toolbox.attr_bool, num_bits)
toolbox.register("population", tools.initRepeat, list, toolbox.individual)
Register the evaluation operator:
toolbox.register("evaluate", eval_func)
Now, register the crossover operator:
toolbox.register("mate", tools.cxTwoPoint)
Register a mutation operator:
toolbox.register("mutate", tools.mutFlipBit, indpb=0.05)
Define the operator for breeding:
toolbox.register("select", tools.selTournament, tournsize=3)
return toolbox
if __name__ == "__main__":
num_bits = 45
toolbox = create_toolbox(num_bits)
random.seed(7)
population = toolbox.population(n=500)
probab_crossing, probab_mutating = 0.5, 0.2
num_generations = 10
print('\nEvolution process starts')
return toolbox
if __name__ == "__main__":
num_bits = 45
toolbox = create_toolbox(num_bits)
random.seed(7)
population = toolbox.population(n=500)
probab_crossing, probab_mutating = 0.5, 0.2
num_generations = 10
print('\nEvolution process starts')
Evaluate the entire population:
fitnesses = list(map(toolbox.evaluate, population))
for ind, fit in zip(population, fitnesses):
ind.fitness.values = fit
print('\nEvaluated', len(population), 'individuals')
for ind, fit in zip(population, fitnesses):
ind.fitness.values = fit
print('\nEvaluated', len(population), 'individuals')
Create and iterate through generations:
for g in range(num_generations):
print("\n- Generation", g)
print("\n- Generation", g)
Selecting the next generation individuals:
offspring = toolbox.select(population, len(population))
Now, clone the selected individuals:
offspring = list(map(toolbox.clone, offspring))
Apply crossover and mutation on the offspring:
for child1, child2 in zip(offspring[::2], offspring[1::2]):
if random.random() < probab_crossing:
toolbox.mate(child1, child2)
if random.random() < probab_crossing:
toolbox.mate(child1, child2)
Delete the fitness value of child:
del child1.fitness.values
del child2.fitness.values
del child2.fitness.values
Now, apply mutation:
for mutant in offspring:
if random.random() < probab_mutating:
toolbox.mutate(mutant)
del mutant.fitness.values
if random.random() < probab_mutating:
toolbox.mutate(mutant)
del mutant.fitness.values
Evaluate the individuals with an invalid fitness:
invalid_ind = [ind for ind in offspring if not ind.fitness.valid]
fitnesses = map(toolbox.evaluate, invalid_ind)
fitnesses = map(toolbox.evaluate, invalid_ind)
for ind, fit in zip(invalid_ind, fitnesses):
ind.fitness.values = fit
print('Evaluated', len(invalid_ind), 'individuals')
ind.fitness.values = fit
print('Evaluated', len(invalid_ind), 'individuals')
Now, replace population with next generation individual:
population[:] = offspring
Print the statistics for the current generations:
fits = [ind.fitness.values[0] for ind in population]
length = len(population)
mean = sum(fits) / length
length = len(population)
mean = sum(fits) / length
sum2 = sum(x*x for x in fits)
std = abs(sum2 / length - mean**2)**0.5
print('Min =', min(fits), ', Max =', max(fits))
print('Average =', round(mean, 2), ', Standard deviation =',
round(std, 2))
print("\n- Evolution ends")
std = abs(sum2 / length - mean**2)**0.5
print('Min =', min(fits), ', Max =', max(fits))
print('Average =', round(mean, 2), ', Standard deviation =',
round(std, 2))
print("\n- Evolution ends")
Print the final output:
best_ind = tools.selBest(population, 1)[0]
print('\nBest individual:\n', best_ind)
print('\nNumber of ones:', sum(best_ind))
print('\nBest individual:\n', best_ind)
print('\nNumber of ones:', sum(best_ind))
Following would be the output:
Evolution process starts
Evaluated 500 individuals
- Generation 0
Evaluated 295 individuals
Min = 32.0 , Max = 45.0
Average = 40.29 , Standard deviation = 2.61
- Generation 1
Evaluated 292 individuals
Min = 34.0 , Max = 45.0
Average = 42.35 , Standard deviation = 1.91
- Generation 2
Evaluated 277 individuals
Min = 37.0 , Max = 45.0
Average = 43.39 , Standard deviation = 1.46
… … … …
- Generation 9
Evaluated 299 individuals
Min = 40.0 , Max = 45.0
Average = 44.12 , Standard deviation = 1.11
- Evolution ends
Best individual:
[0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1]
Number of ones: 15
Evaluated 500 individuals
- Generation 0
Evaluated 295 individuals
Min = 32.0 , Max = 45.0
Average = 40.29 , Standard deviation = 2.61
- Generation 1
Evaluated 292 individuals
Min = 34.0 , Max = 45.0
Average = 42.35 , Standard deviation = 1.91
- Generation 2
Evaluated 277 individuals
Min = 37.0 , Max = 45.0
Average = 43.39 , Standard deviation = 1.46
… … … …
- Generation 9
Evaluated 299 individuals
Min = 40.0 , Max = 45.0
Average = 44.12 , Standard deviation = 1.11
- Evolution ends
Best individual:
[0, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 1, 1, 1, 0, 0, 1, 0, 1]
Number of ones: 15
0 comments:
Post a Comment