Analytics is a collection of techniques and tools used for creating value from data. Techniques include concepts such as artificial intelligence (AI), machine learning (ML), and deep learning (DL) algorithms.
AI, ML, and DL are defined as follows:
1. Artificial Intelligence: Algorithms and systems that exhibit human-like intelligence.
2. Machine Learning: Subset of AI that can learn to perform a task with extracted data and/or models.
3. Deep Learning: Subset of machine learning that imitate the functioning of human brain to solve problems.
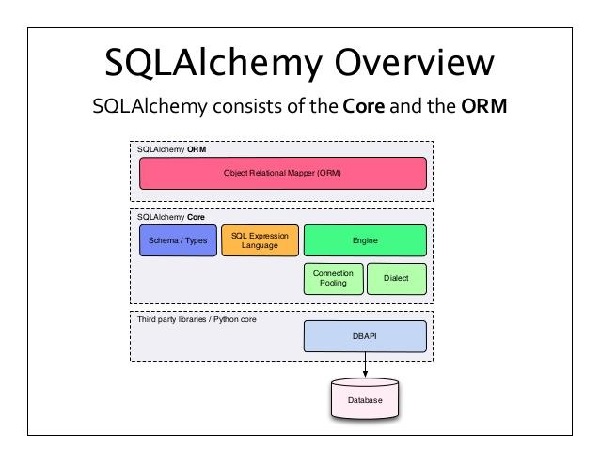
The relationship between AI, ML, and DL can be visualized as shown in Figure below:

There is another school of thought that believes that AI and ML are different (ML is not a subset of AI) with some overlap. The important point is that all of them are algorithms, which are nothing but set of instructions used for solving business and social problems.
Machine learning is a set of algorithms that have the capability to learn to perform tasks such as prediction and classification effectively using data. Learning is achieved using additional data and/or additional models. An algorithm can be called a learning algorithm when it improves on a performance metric while performing a task, for example, accuracy of classification such as fraud, customer churn, and so on. Machine learning algorithms are classified into four categories as defined below:
Supervised Learning Algorithms: These algorithms require the knowledge of both the outcome variable (dependent variable) and the features (independent variable or input variables). The algorithm learns (i.e., estimates the values of the model parameters or feature weights) by defining a loss function which is usually a function of the difference between the predicted value and actual value of the outcome variable. Algorithms such as linear regression, logistic regression, discriminant analysis are examples of supervised learning algorithms. The prediction is achieved (by estimating feature weights) with the knowledge of the actual values of the outcome variables, thus called supervised learning algorithms. That is, the supervision is achieved using the knowledge of outcome variable values.
Unsupervised Learning Algorithms: These algorithms are set of algorithms which do not have the knowledge of the outcome variable in the dataset. The algorithms must find the possible values of the outcome variable. Algorithms such as clustering, principal component analysis are examples of unsupervised learning algorithms. Since the values of outcome variable are unknown in the training data, supervision using that knowledge is not possible.
Reinforcement Learning Algorithms: In many datasets, there could be uncertainty around both input as well as the output variables. For example, consider the case of spell check in various text editors. If a person types “buutiful” in Microsoft Word, the spell check in Microsoft Word will immediately identify this as a spelling mistake and give options such as “beautiful”, “bountiful”, and “dutiful”. Here the prediction is not one single value, but a set of values. Another definition is: Reinforcement learning algorithms are algorithms that have to take sequential actions (decisions) to maximize a cumulative reward. Techniques such as Markov chain and Markov decision process are examples of reinforcement learning algorithms.
Evolutionary Learning Algorithms: Evolutional algorithms are algorithms that imitate natural evolution to solve a problem. Techniques such as genetic algorithm and ant colony optimization fall under the category of evolutionary learning algorithms.
AI, ML, and DL are defined as follows:
1. Artificial Intelligence: Algorithms and systems that exhibit human-like intelligence.
2. Machine Learning: Subset of AI that can learn to perform a task with extracted data and/or models.
3. Deep Learning: Subset of machine learning that imitate the functioning of human brain to solve problems.
The relationship between AI, ML, and DL can be visualized as shown in Figure below:
There is another school of thought that believes that AI and ML are different (ML is not a subset of AI) with some overlap. The important point is that all of them are algorithms, which are nothing but set of instructions used for solving business and social problems.
Machine learning is a set of algorithms that have the capability to learn to perform tasks such as prediction and classification effectively using data. Learning is achieved using additional data and/or additional models. An algorithm can be called a learning algorithm when it improves on a performance metric while performing a task, for example, accuracy of classification such as fraud, customer churn, and so on. Machine learning algorithms are classified into four categories as defined below:
Supervised Learning Algorithms: These algorithms require the knowledge of both the outcome variable (dependent variable) and the features (independent variable or input variables). The algorithm learns (i.e., estimates the values of the model parameters or feature weights) by defining a loss function which is usually a function of the difference between the predicted value and actual value of the outcome variable. Algorithms such as linear regression, logistic regression, discriminant analysis are examples of supervised learning algorithms. The prediction is achieved (by estimating feature weights) with the knowledge of the actual values of the outcome variables, thus called supervised learning algorithms. That is, the supervision is achieved using the knowledge of outcome variable values.
Unsupervised Learning Algorithms: These algorithms are set of algorithms which do not have the knowledge of the outcome variable in the dataset. The algorithms must find the possible values of the outcome variable. Algorithms such as clustering, principal component analysis are examples of unsupervised learning algorithms. Since the values of outcome variable are unknown in the training data, supervision using that knowledge is not possible.
Reinforcement Learning Algorithms: In many datasets, there could be uncertainty around both input as well as the output variables. For example, consider the case of spell check in various text editors. If a person types “buutiful” in Microsoft Word, the spell check in Microsoft Word will immediately identify this as a spelling mistake and give options such as “beautiful”, “bountiful”, and “dutiful”. Here the prediction is not one single value, but a set of values. Another definition is: Reinforcement learning algorithms are algorithms that have to take sequential actions (decisions) to maximize a cumulative reward. Techniques such as Markov chain and Markov decision process are examples of reinforcement learning algorithms.
Evolutionary Learning Algorithms: Evolutional algorithms are algorithms that imitate natural evolution to solve a problem. Techniques such as genetic algorithm and ant colony optimization fall under the category of evolutionary learning algorithms.